restructure readme to match updated template
Browse files- README.md +27 -44
- configs/metadata.json +2 -1
- docs/README.md +27 -44
README.md
CHANGED
|
@@ -8,17 +8,14 @@ license: apache-2.0
|
|
| 8 |
# Model Overview
|
| 9 |
A pre-trained model for volumetric (3D) segmentation of brain tumor subregions from multimodal MRIs based on BraTS 2018 data. The whole pipeline is modified from [clara_pt_brain_mri_segmentation](https://catalog.ngc.nvidia.com/orgs/nvidia/teams/med/models/clara_pt_brain_mri_segmentation).
|
| 10 |
|
| 11 |
-
## Workflow
|
| 12 |
-
|
| 13 |
The model is trained to segment 3 nested subregions of primary brain tumors (gliomas): the "enhancing tumor" (ET), the "tumor core" (TC), the "whole tumor" (WT) based on 4 aligned input MRI scans (T1c, T1, T2, FLAIR).
|
| 14 |
- The ET is described by areas that show hyper intensity in T1c when compared to T1, but also when compared to "healthy" white matter in T1c.
|
| 15 |
- The TC describes the bulk of the tumor, which is what is typically resected. The TC entails the ET, as well as the necrotic (fluid-filled) and the non-enhancing (solid) parts of the tumor.
|
| 16 |
- The WT describes the complete extent of the disease, as it entails the TC and the peritumoral edema (ED), which is typically depicted by hyper-intense signal in FLAIR.
|
| 17 |
|
| 18 |
-

|
| 19 |
|
| 20 |
## Data
|
| 21 |
-
|
| 22 |
The training data is from the [Multimodal Brain Tumor Segmentation Challenge (BraTS) 2018](https://www.med.upenn.edu/cbica/sbia/brats2018/tasks.html).
|
| 23 |
|
| 24 |
- Target: 3 tumor subregions
|
|
@@ -28,16 +25,15 @@ The training data is from the [Multimodal Brain Tumor Segmentation Challenge (Br
|
|
| 28 |
|
| 29 |
The provided labelled data was partitioned, based on our own split, into training (200 studies), validation (42 studies) and testing (43 studies) datasets.
|
| 30 |
|
| 31 |
-
|
|
|
|
| 32 |
|
| 33 |
```
|
| 34 |
python scripts/prepare_datalist.py --path your-brats18-dataset-path
|
| 35 |
```
|
| 36 |
|
| 37 |
## Training configuration
|
| 38 |
-
|
| 39 |
-
This model utilized a similar approach described in 3D MRI brain tumor segmentation
|
| 40 |
-
using autoencoder regularization, which was a winning method in BraTS2018 [1]. The training was performed with the following:
|
| 41 |
|
| 42 |
- GPU: At least 16GB of GPU memory.
|
| 43 |
- Actual Model Input: 224 x 224 x 144
|
|
@@ -47,73 +43,60 @@ using autoencoder regularization, which was a winning method in BraTS2018 [1]. T
|
|
| 47 |
- Loss: DiceLoss
|
| 48 |
|
| 49 |
## Input
|
| 50 |
-
|
| 51 |
-
|
| 52 |
-
|
| 53 |
-
|
| 54 |
-
|
| 55 |
-
3. Randomly spatial flipping
|
| 56 |
-
4. Randomly scaling and shifting intensity of the volume
|
| 57 |
|
| 58 |
## Output
|
| 59 |
-
|
| 60 |
-
Output: 3 channels
|
| 61 |
- Label 0: TC tumor subregion
|
| 62 |
- Label 1: WT tumor subregion
|
| 63 |
- Label 2: ET tumor subregion
|
| 64 |
|
| 65 |
-
##
|
| 66 |
-
|
| 67 |
-
The achieved Dice scores on the validation data are:
|
| 68 |
- Tumor core (TC): 0.8559
|
| 69 |
- Whole tumor (WT): 0.9026
|
| 70 |
- Enhancing tumor (ET): 0.7905
|
| 71 |
- Average: 0.8518
|
| 72 |
|
| 73 |
-
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 74 |
|
| 75 |
-
|
| 76 |
|
|
|
|
| 77 |
```
|
| 78 |
python -m monai.bundle run training --meta_file configs/metadata.json --config_file configs/train.json --logging_file configs/logging.conf
|
| 79 |
```
|
| 80 |
|
| 81 |
-
Override the `train` config to execute multi-GPU training:
|
| 82 |
-
|
| 83 |
```
|
| 84 |
torchrun --standalone --nnodes=1 --nproc_per_node=8 -m monai.bundle run training --meta_file configs/metadata.json --config_file "['configs/train.json','configs/multi_gpu_train.json']" --logging_file configs/logging.conf
|
| 85 |
```
|
| 86 |
|
| 87 |
-
Please note that the distributed training
|
| 88 |
-
Please refer to [pytorch's official tutorial](https://pytorch.org/tutorials/intermediate/ddp_tutorial.html) for more details.
|
| 89 |
-
|
| 90 |
-
Override the `train` config to execute evaluation with the trained model:
|
| 91 |
|
|
|
|
| 92 |
```
|
| 93 |
python -m monai.bundle run evaluating --meta_file configs/metadata.json --config_file "['configs/train.json','configs/evaluate.json']" --logging_file configs/logging.conf
|
| 94 |
```
|
| 95 |
|
| 96 |
-
Execute inference:
|
| 97 |
-
|
| 98 |
```
|
| 99 |
python -m monai.bundle run evaluating --meta_file configs/metadata.json --config_file configs/inference.json --logging_file configs/logging.conf
|
| 100 |
```
|
| 101 |
|
| 102 |
-
# Training
|
| 103 |
-
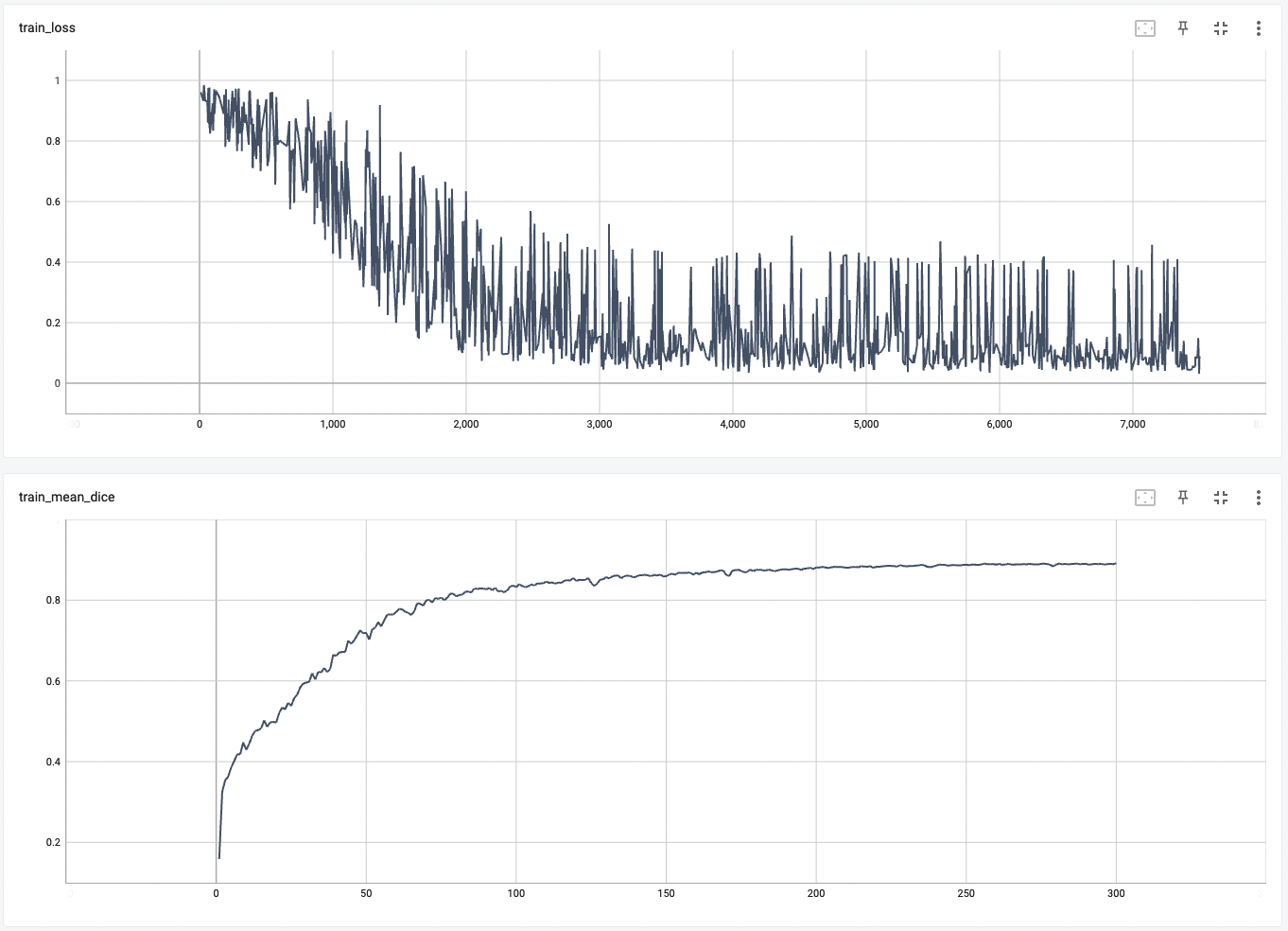
A graph showing the training loss and the mean dice over 300 epochs.
|
| 104 |
-
|
| 105 |
-
|
| 106 |
-
# Validation
|
| 107 |
-
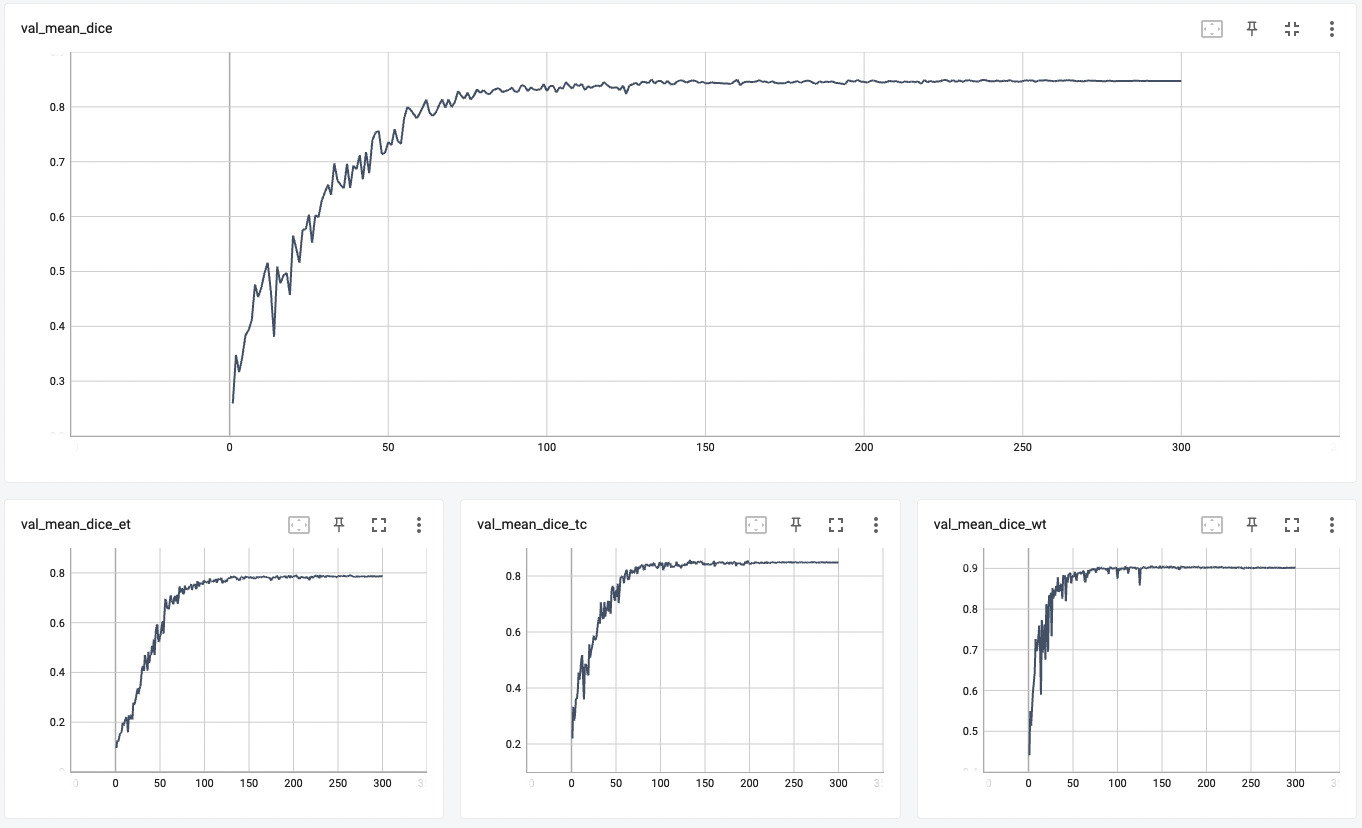
A graph showing the validation mean dice over 300 epochs.
|
| 108 |
-
|
| 109 |
-
|
| 110 |
-
|
| 111 |
-
# Disclaimer
|
| 112 |
-
|
| 113 |
-
This is an example, not to be used for diagnostic purposes.
|
| 114 |
-
|
| 115 |
# References
|
| 116 |
-
|
| 117 |
[1] Myronenko, Andriy. "3D MRI brain tumor segmentation using autoencoder regularization." International MICCAI Brainlesion Workshop. Springer, Cham, 2018. https://arxiv.org/abs/1810.11654.
|
| 118 |
|
| 119 |
# License
|
|
|
|
| 8 |
# Model Overview
|
| 9 |
A pre-trained model for volumetric (3D) segmentation of brain tumor subregions from multimodal MRIs based on BraTS 2018 data. The whole pipeline is modified from [clara_pt_brain_mri_segmentation](https://catalog.ngc.nvidia.com/orgs/nvidia/teams/med/models/clara_pt_brain_mri_segmentation).
|
| 10 |
|
|
|
|
|
|
|
| 11 |
The model is trained to segment 3 nested subregions of primary brain tumors (gliomas): the "enhancing tumor" (ET), the "tumor core" (TC), the "whole tumor" (WT) based on 4 aligned input MRI scans (T1c, T1, T2, FLAIR).
|
| 12 |
- The ET is described by areas that show hyper intensity in T1c when compared to T1, but also when compared to "healthy" white matter in T1c.
|
| 13 |
- The TC describes the bulk of the tumor, which is what is typically resected. The TC entails the ET, as well as the necrotic (fluid-filled) and the non-enhancing (solid) parts of the tumor.
|
| 14 |
- The WT describes the complete extent of the disease, as it entails the TC and the peritumoral edema (ED), which is typically depicted by hyper-intense signal in FLAIR.
|
| 15 |
|
| 16 |
+

|
| 17 |
|
| 18 |
## Data
|
|
|
|
| 19 |
The training data is from the [Multimodal Brain Tumor Segmentation Challenge (BraTS) 2018](https://www.med.upenn.edu/cbica/sbia/brats2018/tasks.html).
|
| 20 |
|
| 21 |
- Target: 3 tumor subregions
|
|
|
|
| 25 |
|
| 26 |
The provided labelled data was partitioned, based on our own split, into training (200 studies), validation (42 studies) and testing (43 studies) datasets.
|
| 27 |
|
| 28 |
+
### Preprocessing
|
| 29 |
+
The data list/split can be created with the script `scripts/prepare_datalist.py`.
|
| 30 |
|
| 31 |
```
|
| 32 |
python scripts/prepare_datalist.py --path your-brats18-dataset-path
|
| 33 |
```
|
| 34 |
|
| 35 |
## Training configuration
|
| 36 |
+
This model utilized a similar approach described in 3D MRI brain tumor segmentation using autoencoder regularization, which was a winning method in BraTS2018 [1]. The training was performed with the following:
|
|
|
|
|
|
|
| 37 |
|
| 38 |
- GPU: At least 16GB of GPU memory.
|
| 39 |
- Actual Model Input: 224 x 224 x 144
|
|
|
|
| 43 |
- Loss: DiceLoss
|
| 44 |
|
| 45 |
## Input
|
| 46 |
+
4 channel aligned MRIs at 1 x 1 x 1 mm
|
| 47 |
+
- T1c
|
| 48 |
+
- T1
|
| 49 |
+
- T2
|
| 50 |
+
- FLAIR
|
|
|
|
|
|
|
| 51 |
|
| 52 |
## Output
|
| 53 |
+
3 channels
|
|
|
|
| 54 |
- Label 0: TC tumor subregion
|
| 55 |
- Label 1: WT tumor subregion
|
| 56 |
- Label 2: ET tumor subregion
|
| 57 |
|
| 58 |
+
## Performance
|
| 59 |
+
Dice score was used for evaluating the performance of the model. This model achieved Dice scores on the validation data of:
|
|
|
|
| 60 |
- Tumor core (TC): 0.8559
|
| 61 |
- Whole tumor (WT): 0.9026
|
| 62 |
- Enhancing tumor (ET): 0.7905
|
| 63 |
- Average: 0.8518
|
| 64 |
|
| 65 |
+
#### Training Loss and Dice
|
| 66 |
+

|
| 67 |
+
|
| 68 |
+
#### Validation Dice
|
| 69 |
+
|
| 70 |
+

|
| 71 |
+
|
| 72 |
+
## MONAI Bundle Commands
|
| 73 |
+
In addition to the Pythonic APIs, a few command line interfaces (CLI) are provided to interact with the bundle. The CLI supports flexible use cases, such as overriding configs at runtime and predefining arguments in a file.
|
| 74 |
|
| 75 |
+
For more details usage instructions, visit the [MONAI Bundle Configuration Page](https://docs.monai.io/en/latest/config_syntax.html).
|
| 76 |
|
| 77 |
+
#### Execute training:
|
| 78 |
```
|
| 79 |
python -m monai.bundle run training --meta_file configs/metadata.json --config_file configs/train.json --logging_file configs/logging.conf
|
| 80 |
```
|
| 81 |
|
| 82 |
+
#### Override the `train` config to execute multi-GPU training:
|
|
|
|
| 83 |
```
|
| 84 |
torchrun --standalone --nnodes=1 --nproc_per_node=8 -m monai.bundle run training --meta_file configs/metadata.json --config_file "['configs/train.json','configs/multi_gpu_train.json']" --logging_file configs/logging.conf
|
| 85 |
```
|
| 86 |
|
| 87 |
+
Please note that the distributed training-related options depend on the actual running environment; thus, users may need to remove `--standalone`, modify `--nnodes`, or do some other necessary changes according to the machine used. For more details, please refer to [pytorch's official tutorial](https://pytorch.org/tutorials/intermediate/ddp_tutorial.html).
|
|
|
|
|
|
|
|
|
|
| 88 |
|
| 89 |
+
#### Override the `train` config to execute evaluation with the trained model:
|
| 90 |
```
|
| 91 |
python -m monai.bundle run evaluating --meta_file configs/metadata.json --config_file "['configs/train.json','configs/evaluate.json']" --logging_file configs/logging.conf
|
| 92 |
```
|
| 93 |
|
| 94 |
+
#### Execute inference:
|
|
|
|
| 95 |
```
|
| 96 |
python -m monai.bundle run evaluating --meta_file configs/metadata.json --config_file configs/inference.json --logging_file configs/logging.conf
|
| 97 |
```
|
| 98 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 99 |
# References
|
|
|
|
| 100 |
[1] Myronenko, Andriy. "3D MRI brain tumor segmentation using autoencoder regularization." International MICCAI Brainlesion Workshop. Springer, Cham, 2018. https://arxiv.org/abs/1810.11654.
|
| 101 |
|
| 102 |
# License
|
configs/metadata.json
CHANGED
|
@@ -1,7 +1,8 @@
|
|
| 1 |
{
|
| 2 |
"schema": "https://github.com/Project-MONAI/MONAI-extra-test-data/releases/download/0.8.1/meta_schema_20220324.json",
|
| 3 |
-
"version": "0.3.
|
| 4 |
"changelog": {
|
|
|
|
| 5 |
"0.3.6": "added train/val graphs",
|
| 6 |
"0.3.5": "update prepare datalist function",
|
| 7 |
"0.3.4": "update output format of inference",
|
|
|
|
| 1 |
{
|
| 2 |
"schema": "https://github.com/Project-MONAI/MONAI-extra-test-data/releases/download/0.8.1/meta_schema_20220324.json",
|
| 3 |
+
"version": "0.3.7",
|
| 4 |
"changelog": {
|
| 5 |
+
"0.3.7": "restructure readme to match updated template",
|
| 6 |
"0.3.6": "added train/val graphs",
|
| 7 |
"0.3.5": "update prepare datalist function",
|
| 8 |
"0.3.4": "update output format of inference",
|
docs/README.md
CHANGED
|
@@ -1,17 +1,14 @@
|
|
| 1 |
# Model Overview
|
| 2 |
A pre-trained model for volumetric (3D) segmentation of brain tumor subregions from multimodal MRIs based on BraTS 2018 data. The whole pipeline is modified from [clara_pt_brain_mri_segmentation](https://catalog.ngc.nvidia.com/orgs/nvidia/teams/med/models/clara_pt_brain_mri_segmentation).
|
| 3 |
|
| 4 |
-
## Workflow
|
| 5 |
-
|
| 6 |
The model is trained to segment 3 nested subregions of primary brain tumors (gliomas): the "enhancing tumor" (ET), the "tumor core" (TC), the "whole tumor" (WT) based on 4 aligned input MRI scans (T1c, T1, T2, FLAIR).
|
| 7 |
- The ET is described by areas that show hyper intensity in T1c when compared to T1, but also when compared to "healthy" white matter in T1c.
|
| 8 |
- The TC describes the bulk of the tumor, which is what is typically resected. The TC entails the ET, as well as the necrotic (fluid-filled) and the non-enhancing (solid) parts of the tumor.
|
| 9 |
- The WT describes the complete extent of the disease, as it entails the TC and the peritumoral edema (ED), which is typically depicted by hyper-intense signal in FLAIR.
|
| 10 |
|
| 11 |
-

|
| 12 |
|
| 13 |
## Data
|
| 14 |
-
|
| 15 |
The training data is from the [Multimodal Brain Tumor Segmentation Challenge (BraTS) 2018](https://www.med.upenn.edu/cbica/sbia/brats2018/tasks.html).
|
| 16 |
|
| 17 |
- Target: 3 tumor subregions
|
|
@@ -21,16 +18,15 @@ The training data is from the [Multimodal Brain Tumor Segmentation Challenge (Br
|
|
| 21 |
|
| 22 |
The provided labelled data was partitioned, based on our own split, into training (200 studies), validation (42 studies) and testing (43 studies) datasets.
|
| 23 |
|
| 24 |
-
|
|
|
|
| 25 |
|
| 26 |
```
|
| 27 |
python scripts/prepare_datalist.py --path your-brats18-dataset-path
|
| 28 |
```
|
| 29 |
|
| 30 |
## Training configuration
|
| 31 |
-
|
| 32 |
-
This model utilized a similar approach described in 3D MRI brain tumor segmentation
|
| 33 |
-
using autoencoder regularization, which was a winning method in BraTS2018 [1]. The training was performed with the following:
|
| 34 |
|
| 35 |
- GPU: At least 16GB of GPU memory.
|
| 36 |
- Actual Model Input: 224 x 224 x 144
|
|
@@ -40,73 +36,60 @@ using autoencoder regularization, which was a winning method in BraTS2018 [1]. T
|
|
| 40 |
- Loss: DiceLoss
|
| 41 |
|
| 42 |
## Input
|
| 43 |
-
|
| 44 |
-
|
| 45 |
-
|
| 46 |
-
|
| 47 |
-
|
| 48 |
-
3. Randomly spatial flipping
|
| 49 |
-
4. Randomly scaling and shifting intensity of the volume
|
| 50 |
|
| 51 |
## Output
|
| 52 |
-
|
| 53 |
-
Output: 3 channels
|
| 54 |
- Label 0: TC tumor subregion
|
| 55 |
- Label 1: WT tumor subregion
|
| 56 |
- Label 2: ET tumor subregion
|
| 57 |
|
| 58 |
-
##
|
| 59 |
-
|
| 60 |
-
The achieved Dice scores on the validation data are:
|
| 61 |
- Tumor core (TC): 0.8559
|
| 62 |
- Whole tumor (WT): 0.9026
|
| 63 |
- Enhancing tumor (ET): 0.7905
|
| 64 |
- Average: 0.8518
|
| 65 |
|
| 66 |
-
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 67 |
|
| 68 |
-
|
| 69 |
|
|
|
|
| 70 |
```
|
| 71 |
python -m monai.bundle run training --meta_file configs/metadata.json --config_file configs/train.json --logging_file configs/logging.conf
|
| 72 |
```
|
| 73 |
|
| 74 |
-
Override the `train` config to execute multi-GPU training:
|
| 75 |
-
|
| 76 |
```
|
| 77 |
torchrun --standalone --nnodes=1 --nproc_per_node=8 -m monai.bundle run training --meta_file configs/metadata.json --config_file "['configs/train.json','configs/multi_gpu_train.json']" --logging_file configs/logging.conf
|
| 78 |
```
|
| 79 |
|
| 80 |
-
Please note that the distributed training
|
| 81 |
-
Please refer to [pytorch's official tutorial](https://pytorch.org/tutorials/intermediate/ddp_tutorial.html) for more details.
|
| 82 |
-
|
| 83 |
-
Override the `train` config to execute evaluation with the trained model:
|
| 84 |
|
|
|
|
| 85 |
```
|
| 86 |
python -m monai.bundle run evaluating --meta_file configs/metadata.json --config_file "['configs/train.json','configs/evaluate.json']" --logging_file configs/logging.conf
|
| 87 |
```
|
| 88 |
|
| 89 |
-
Execute inference:
|
| 90 |
-
|
| 91 |
```
|
| 92 |
python -m monai.bundle run evaluating --meta_file configs/metadata.json --config_file configs/inference.json --logging_file configs/logging.conf
|
| 93 |
```
|
| 94 |
|
| 95 |
-
# Training
|
| 96 |
-
A graph showing the training loss and the mean dice over 300 epochs.
|
| 97 |
-

|
| 98 |
-
|
| 99 |
-
# Validation
|
| 100 |
-
A graph showing the validation mean dice over 300 epochs.
|
| 101 |
-

|
| 102 |
-
|
| 103 |
-
|
| 104 |
-
# Disclaimer
|
| 105 |
-
|
| 106 |
-
This is an example, not to be used for diagnostic purposes.
|
| 107 |
-
|
| 108 |
# References
|
| 109 |
-
|
| 110 |
[1] Myronenko, Andriy. "3D MRI brain tumor segmentation using autoencoder regularization." International MICCAI Brainlesion Workshop. Springer, Cham, 2018. https://arxiv.org/abs/1810.11654.
|
| 111 |
|
| 112 |
# License
|
|
|
|
| 1 |
# Model Overview
|
| 2 |
A pre-trained model for volumetric (3D) segmentation of brain tumor subregions from multimodal MRIs based on BraTS 2018 data. The whole pipeline is modified from [clara_pt_brain_mri_segmentation](https://catalog.ngc.nvidia.com/orgs/nvidia/teams/med/models/clara_pt_brain_mri_segmentation).
|
| 3 |
|
|
|
|
|
|
|
| 4 |
The model is trained to segment 3 nested subregions of primary brain tumors (gliomas): the "enhancing tumor" (ET), the "tumor core" (TC), the "whole tumor" (WT) based on 4 aligned input MRI scans (T1c, T1, T2, FLAIR).
|
| 5 |
- The ET is described by areas that show hyper intensity in T1c when compared to T1, but also when compared to "healthy" white matter in T1c.
|
| 6 |
- The TC describes the bulk of the tumor, which is what is typically resected. The TC entails the ET, as well as the necrotic (fluid-filled) and the non-enhancing (solid) parts of the tumor.
|
| 7 |
- The WT describes the complete extent of the disease, as it entails the TC and the peritumoral edema (ED), which is typically depicted by hyper-intense signal in FLAIR.
|
| 8 |
|
| 9 |
+

|
| 10 |
|
| 11 |
## Data
|
|
|
|
| 12 |
The training data is from the [Multimodal Brain Tumor Segmentation Challenge (BraTS) 2018](https://www.med.upenn.edu/cbica/sbia/brats2018/tasks.html).
|
| 13 |
|
| 14 |
- Target: 3 tumor subregions
|
|
|
|
| 18 |
|
| 19 |
The provided labelled data was partitioned, based on our own split, into training (200 studies), validation (42 studies) and testing (43 studies) datasets.
|
| 20 |
|
| 21 |
+
### Preprocessing
|
| 22 |
+
The data list/split can be created with the script `scripts/prepare_datalist.py`.
|
| 23 |
|
| 24 |
```
|
| 25 |
python scripts/prepare_datalist.py --path your-brats18-dataset-path
|
| 26 |
```
|
| 27 |
|
| 28 |
## Training configuration
|
| 29 |
+
This model utilized a similar approach described in 3D MRI brain tumor segmentation using autoencoder regularization, which was a winning method in BraTS2018 [1]. The training was performed with the following:
|
|
|
|
|
|
|
| 30 |
|
| 31 |
- GPU: At least 16GB of GPU memory.
|
| 32 |
- Actual Model Input: 224 x 224 x 144
|
|
|
|
| 36 |
- Loss: DiceLoss
|
| 37 |
|
| 38 |
## Input
|
| 39 |
+
4 channel aligned MRIs at 1 x 1 x 1 mm
|
| 40 |
+
- T1c
|
| 41 |
+
- T1
|
| 42 |
+
- T2
|
| 43 |
+
- FLAIR
|
|
|
|
|
|
|
| 44 |
|
| 45 |
## Output
|
| 46 |
+
3 channels
|
|
|
|
| 47 |
- Label 0: TC tumor subregion
|
| 48 |
- Label 1: WT tumor subregion
|
| 49 |
- Label 2: ET tumor subregion
|
| 50 |
|
| 51 |
+
## Performance
|
| 52 |
+
Dice score was used for evaluating the performance of the model. This model achieved Dice scores on the validation data of:
|
|
|
|
| 53 |
- Tumor core (TC): 0.8559
|
| 54 |
- Whole tumor (WT): 0.9026
|
| 55 |
- Enhancing tumor (ET): 0.7905
|
| 56 |
- Average: 0.8518
|
| 57 |
|
| 58 |
+
#### Training Loss and Dice
|
| 59 |
+

|
| 60 |
+
|
| 61 |
+
#### Validation Dice
|
| 62 |
+
|
| 63 |
+

|
| 64 |
+
|
| 65 |
+
## MONAI Bundle Commands
|
| 66 |
+
In addition to the Pythonic APIs, a few command line interfaces (CLI) are provided to interact with the bundle. The CLI supports flexible use cases, such as overriding configs at runtime and predefining arguments in a file.
|
| 67 |
|
| 68 |
+
For more details usage instructions, visit the [MONAI Bundle Configuration Page](https://docs.monai.io/en/latest/config_syntax.html).
|
| 69 |
|
| 70 |
+
#### Execute training:
|
| 71 |
```
|
| 72 |
python -m monai.bundle run training --meta_file configs/metadata.json --config_file configs/train.json --logging_file configs/logging.conf
|
| 73 |
```
|
| 74 |
|
| 75 |
+
#### Override the `train` config to execute multi-GPU training:
|
|
|
|
| 76 |
```
|
| 77 |
torchrun --standalone --nnodes=1 --nproc_per_node=8 -m monai.bundle run training --meta_file configs/metadata.json --config_file "['configs/train.json','configs/multi_gpu_train.json']" --logging_file configs/logging.conf
|
| 78 |
```
|
| 79 |
|
| 80 |
+
Please note that the distributed training-related options depend on the actual running environment; thus, users may need to remove `--standalone`, modify `--nnodes`, or do some other necessary changes according to the machine used. For more details, please refer to [pytorch's official tutorial](https://pytorch.org/tutorials/intermediate/ddp_tutorial.html).
|
|
|
|
|
|
|
|
|
|
| 81 |
|
| 82 |
+
#### Override the `train` config to execute evaluation with the trained model:
|
| 83 |
```
|
| 84 |
python -m monai.bundle run evaluating --meta_file configs/metadata.json --config_file "['configs/train.json','configs/evaluate.json']" --logging_file configs/logging.conf
|
| 85 |
```
|
| 86 |
|
| 87 |
+
#### Execute inference:
|
|
|
|
| 88 |
```
|
| 89 |
python -m monai.bundle run evaluating --meta_file configs/metadata.json --config_file configs/inference.json --logging_file configs/logging.conf
|
| 90 |
```
|
| 91 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 92 |
# References
|
|
|
|
| 93 |
[1] Myronenko, Andriy. "3D MRI brain tumor segmentation using autoencoder regularization." International MICCAI Brainlesion Workshop. Springer, Cham, 2018. https://arxiv.org/abs/1810.11654.
|
| 94 |
|
| 95 |
# License
|