Datasets:
language:
- en
license: cc-by-4.0
tags:
- physics
task_categories:
- time-series-forecasting
- other
task_ids:
- multivariate-time-series-forecasting
This Dataset is part of The Well Collection.
How To Load from HuggingFace Hub
- Be sure to have
the_wellinstalled (pip install the_well) - Use the
WellDataModuleto retrieve data as follows:
from the_well.data import WellDataModule
# The following line may take a couple of minutes to instantiate the datamodule
datamodule = WellDataModule(
"hf://datasets/polymathic-ai/",
"gray_scott_reaction_diffusion",
)
train_dataloader = datamodule.train_dataloader()
for batch in dataloader:
# Process training batch
...
Pattern formation in the Gray-Scott reaction-diffusion equations
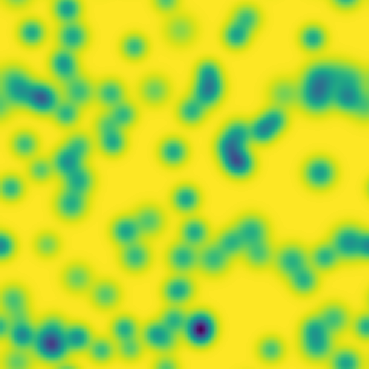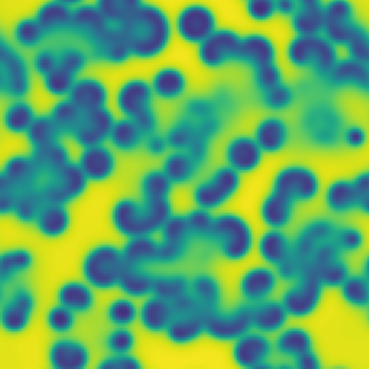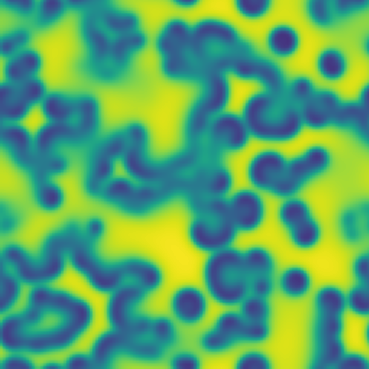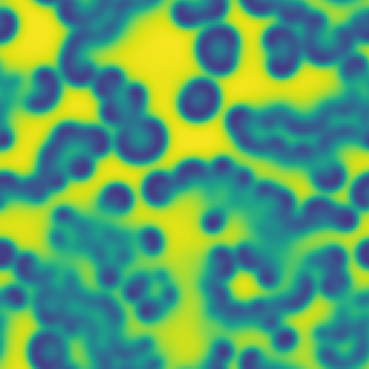
One line description of the data: Stable Turing patterns emerge from randomness, with drastic qualitative differences in pattern dynamics depending on the equation parameters.
Longer description of the data: The Gray-Scott equations are a set of coupled reaction-diffusion equations describing two chemical species, and , whose concentrations vary in space and time. The two parameters and control the “feed” and “kill” rates in the reaction. A zoo of qualitatively different static and dynamic patterns in the solutions are possible depending on these two parameters. There is a rich landscape of pattern formation hidden in these equations.
Associated paper: None.
Domain expert: Daniel Fortunato, CCM and CCB, Flatiron Institute.
Code or software used to generate the data: Github repository (MATLAB R2023a, using the stiff PDE integrator implemented in Chebfun. The Fourier spectral method is used in space (with nonlinear terms evaluated pseudospectrally), and the exponential time-differencing fourth-order Runge-Kutta scheme (ETDRK4) is used in time.)
Equation describing the data
The dimensionless parameters describing the behavior are: the rate at which is replenished (feed rate), the rate at which is removed from the system, and the diffusion coefficients of both species.
| Dataset | FNO | TFNO | Unet | CNextU-net |
|---|---|---|---|---|
gray_scott_reaction_diffusion |
0.3633 | 0.2252 | \(\mathbf{0.1761}\) |
Table: VRMSE metrics on test sets (lower is better). Best results are shown in bold. VRMSE is scaled such that predicting the mean value of the target field results in a score of 1.
About the data
Dimension of discretized data: 1001 time-steps of 128 128 images.
Fields available in the data: The concentration of two chemical species and .
Number of trajectories: 6 sets of parameters, 200 initial conditions per set = 1200.
Estimated size of the ensemble of all simulations: 153.8 GB.
Grid type: uniform, cartesian coordinates.
Initial conditions: Two types of initial conditions generated: random Fourier series and random clusters of Gaussians.
Boundary conditions: periodic.
Simulation time-step: 1 second.
Data are stored separated by ( ): 10 seconds.
Total time range ( to ): , .
Spatial domain size ( , ): .
Set of coefficients or non-dimensional parameters evaluated: All simulations used and . "Gliders": . "Bubbles": . "Maze": . "Worms": . "Spirals": . "Spots": .
Approximate time to generate the data: 5.5 hours per set of parameters, 33 hours total.
Hardware used to generate the data: 40 CPU cores.
What is interesting and challenging about the data:
What phenomena of physical interest are catpured in the data: Pattern formation: by sweeping the two parameters and , a multitude of steady and dynamic patterns can form from random initial conditions.
How to evaluate a new simulator operating in this space: It would be impressive if a simulator—trained only on some of the patterns produced by a subset of the parameter space—could perform well on an unseen set of parameter values that produce fundamentally different patterns. Stability for steady-state patterns over long rollout times would also be impressive.
Warning: Due to the nature of the problem and the possibility to reach an equilibrium for certain values of the kill and feed parameters, a constant stationary behavior can be reached. Here are the trajectories for which a stationary behavior was identified for specy as well as the corresponding time at which it was reached:
Validation set:
- :
- Trajectory 7, time = 123
- Trajectory 8, time = 125
- Trajectory 10, time = 123
- Trajectory 11, time = 125
- Trajectory 12, time = 121
- Trajectory 14, time = 121
- Trajectory 15, time = 129
- Trajectory 16, time = 124
- Trajectory 17, time = 122
- Trajectory 18, time = 121
- Trajectory 19, time = 155
- :
- Trajectory 14, time = 109
- :
Training set:
- :
- Trajectory 81, time = 126
- Trajectory 82, time = 126
- Trajectory 83, time = 123
- Trajectory 85, time = 123
- Trajectory 86, time = 124
- Trajectory 87, time = 127
- Trajectory 88, time = 121
- Trajectory 90, time = 123
- Trajectory 91, time = 121
- Trajectory 92, time = 126
- Trajectory 93, time = 121
- Trajectory 94, time = 126
- Trajectory 95, time = 125
- Trajectory 96, time = 123
- Trajectory 97, time = 126
- Trajectory 98, time = 121
- Trajectory 99, time = 125
- Trajectory 100, time = 126
- Trajectory 101, time = 125
- Trajectory 102, time = 159
- Trajectory 103, time = 129
- Trajectory 105, time = 125
- Trajectory 107, time = 122
- Trajectory 108, time = 126
- Trajectory 110, time = 127
- Trajectory 111, time = 122
- Trajectory 112, time = 121
- Trajectory 113, time = 122
- Trajectory 114, time = 126
- Trajectory 115, time = 126
- Trajectory 116, time = 126
- Trajectory 117, time = 122
- Trajectory 118, time = 123
- Trajectory 119, time = 123
- Trajectory 120, time = 125
- Trajectory 121, time = 126
- Trajectory 122, time = 121
- Trajectory 123, time = 122
- Trajectory 125, time = 125
- Trajectory 126, time = 127
- Trajectory 127, time = 125
- Trajectory 129, time = 125
- Trajectory 130, time = 122
- Trajectory 131, time = 125
- Trajectory 132, time = 131
- Trajectory 133, time = 126
- Trajectory 134, time = 159
- Trajectory 135, time = 121
- Trajectory 136, time = 126
- Trajectory 137, time = 125
- Trajectory 138, time = 126
- Trajectory 139, time = 123
- Trajectory 140, time = 128
- Trajectory 141, time = 126
- Trajectory 142, time = 123
- Trajectory 144, time = 122
- Trajectory 145, time = 125
- Trajectory 146, time = 123
- Trajectory 147, time = 126
- Trajectory 148, time = 121
- Trajectory 149, time = 122
- Trajectory 150, time = 125
- Trajectory 151, time = 126
- Trajectory 152, time = 152
- Trajectory 153, time = 127
- Trajectory 154, time = 122
- Trajectory 155, time = 124
- Trajectory 156, time = 122
- Trajectory 158, time = 126
- Trajectory 159, time = 121
- :
- Trajectory 97, time = 109
- Trajectory 134, time = 107
- Trajectory 147, time = 109
- Trajectory 153, time = 112
- :
Test set:
- :
- Trajectory 12, time = 127
- Trajectory 13, time = 125
- Trajectory 14, time = 123
- Trajectory 15, time = 126
- Trajectory 16, time = 126
- Trajectory 17, time = 123
- Trajectory 18, time = 128
- Trajectory 19, time = 125
- :
- Trajectory 11, time = 113
- :