license: other
AIDO.StructurePrediction






.png)
.png)
Model Description
AIDO.StructurePrediction is an AlphaFold3-like full-atom structure prediction model, designed to predict the structure and interactions of biological molecules, including proteins, DNA, RNA, ligands, and antibodies. This model harnesses both structural and sequence modalities to provide high-fidelity predictions for various biological tasks. Our model achieved state-of-the-art performance on immunology-related structure prediction tasks, including antibody, nanobody, antibody-antigen, and nanobody-antigen.
Model Details
Key Features
- Multi-Modal Learning: Combines 3D structural and sequence data (nucleotides and amino acids) to enhance model accuracy and applicability.
- High-Quality Data: We have used carefully curated structure data when training the model.
- Data Augmentation: Implements novel data augmentation and distillation techniques to diversify training datasets, improving robustness and generalization.
- Integration of Multiple Sequence Alignments (MSA): Utilizes alignment data from diverse biological databases to improve predictive capabilities.
- Training Strategies: Incorporates advanced training methodologies to refine model performance and efficiency.
Model Architecture
- Type: Pairformer+Diffusion model architecture.
- Key Components:
- Pairformer: Designed to learn complex relationships from both single sequences and multiple sequence alignments.
- Diffusion Module: Generates multiple conformations of the structure.
- Hyperparameters:
- Some key parameters:
Model Arch Component Value Pairformer Blocks 48 MSA Moduel Blocks 4 Diffusion Module Blocks 24 Diffusion Heads 16 - Hyperparameters can be found in inference_v0.1.yaml
- Some key parameters:
Usage
Please see experiments/AIDO.StructurePrediction in AIDO.ModelGenerator for more details.
Model Performance
Model Evaluation Metrics
RMSD: Root Mean Square Deviation between prediction and ground truth.
- Protein/Antibody: We calculate the RMSD for Cα atoms.
- DNA/RNA: We calculate the RMSD for C1 atoms.
- Ligand: We use the coordinates of all atoms.
When calculating RMSD for protein-ligand, RNA-ligand, and DNA-ligand interactions, if we use only Cα and C1 for proteins, RNAs, and DNAs, while using full atom coordinates for ligands, the metric may be affected by the number of atoms in the ligand. This could create potential issues. We plan to address this problem in the future.
DockQ:
We modified the script based on this public repo to support missing residues.
Note: For all the metrics mentioned above, if there are missing residues or atoms, we will input the complete information into our model. Because the ground truth structure doesn't include the coordinates of these components, evaluating this type of data can be very challenging. Fortunately, we know exactly which residues or atoms are missing, so we do not need to use any approximated alignment when calculating these metrics. We have found that using approximated alignments in metric calculations can sometimes result in inaccurate metric values and hinder head-to-head comparisons between different methods.
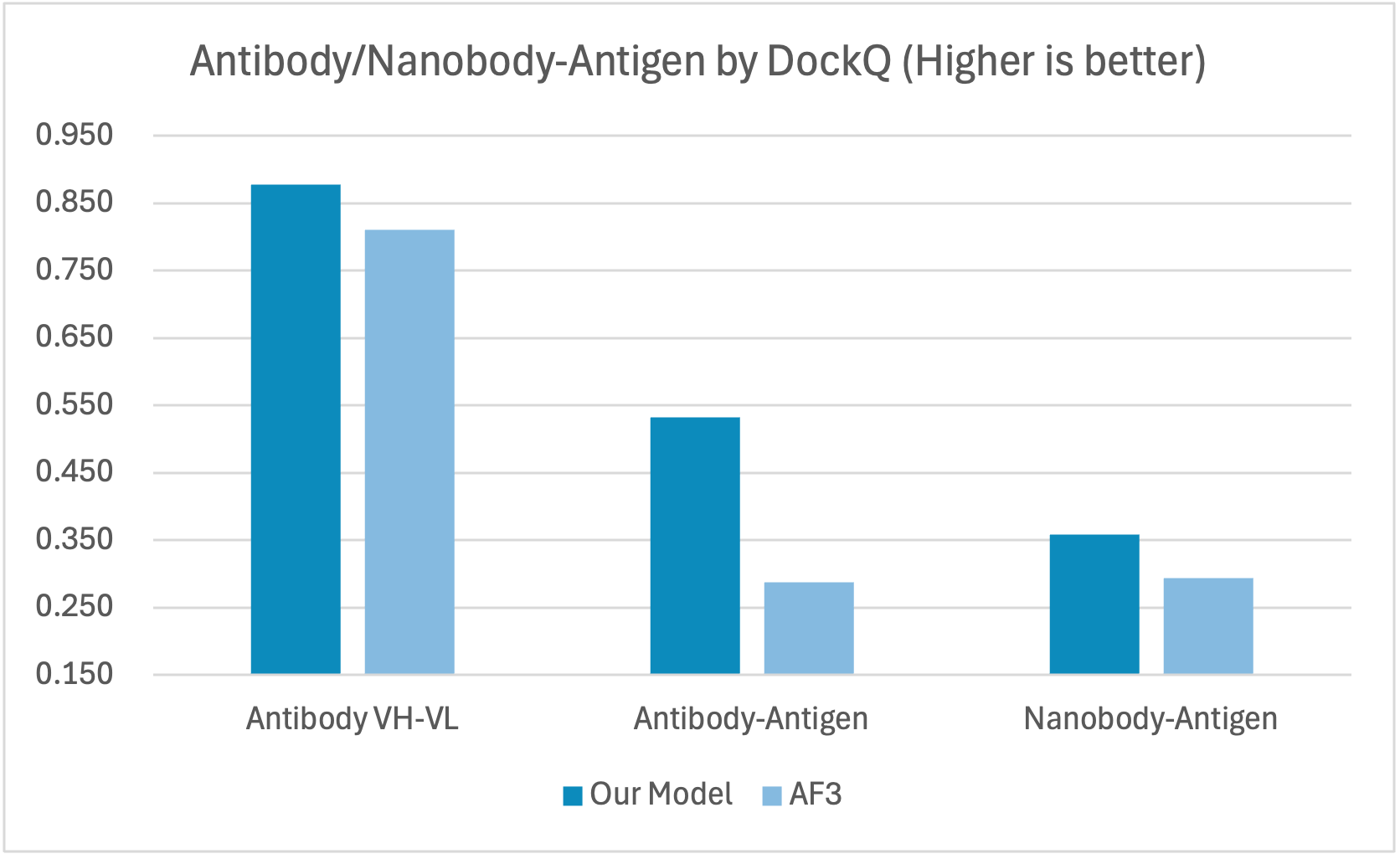
Performance
The antibody/nanobody-antigen data used for the evaluation was curated from recently released PDBs after September 30, 2021. We also assessed the quality of the selected structures to ensure the interfaces are valid. For instance, the binding sites are typically located in the complementarity-determining regions (CDRs) for antibody-antigen or nanobody-antigen complexes. Additionally, we examined the distance map between the heavy chain and light chain to confirm that the selected chain pair constitutes a valid antibody.

License and Disclaimer
Unless otherwise stated, this project is licensed under the GenBio AI Community License Agreement. This project includes third-party components (MMseqs, Protenix). Use of this project does not override or waive the original license terms of these third-party components - you are still bound by their respective licenses and can download from their original sites.
Citation
Please cite AIDO.StructurePrediction using the following BibTex code:
@inproceedings{aido_structurepediction,
title = {AIDO StructurePrediction},
url = {https://huggingface.co/genbio-ai/AIDO.StructurePrediction},
author = {Kun Leo, Jiayou Zhang, Georgy Andreev, Hugo Ly, Le Song, Eric P Xing},
year = {2025},
}