Mejurix Medical-Legal Embedding Model
This model is a fine-tuned Transformer (BERT-based) that generates high-quality embeddings for documents in medical and legal domains, with a focus on capturing the semantic relationships between medical and legal concepts. The model leverages NER (Named Entity Recognition) to better understand domain-specific entities and their relationships.
Model Description
Model Architecture
- Base Architecture: BERT (Bidirectional Encoder Representations from Transformers)
- Base Model: medicalai/ClinicalBERT
- Modifications:
- Custom embedding projection layer (768 → 256 dimensions)
- NER-enhanced attention mechanism
- Domain-specific fine-tuning
Key Features
- Domain-Specific Embeddings: Optimized for medical and legal text analysis
- NER-Enhanced Understanding: Utilizes named entity recognition to improve context awareness
- Reduced Dimensionality: 256-dimensional embeddings balance expressiveness and efficiency
- Cross-Domain Connections: Effectively captures relationships between medical findings and legal implications
- Transformer-Based: Leverages bidirectional attention mechanisms for better context understanding
Performance Comparison
Our model outperforms other similar domain-specific models:
| Model | Avg Similarity | #Params | Notes |
|---|---|---|---|
| Mejurix (ours) | 0.9859 | 110M | Medical-legal + NER FT |
| ClinicalBERT | 0.9719 | 110M | No NER, no fine-tuning |
| BioBERT | 0.9640 | 110M | Domain medical only |
| LegalBERT | 0.9508 | 110M | Domain legal only |
The Mejurix model shows superior performance across all relationship types, particularly in cross-domain relationships between medical and legal concepts.
Detailed Relationship-Type Comparison
Our model demonstrates consistently higher similarity scores across all relationship types compared to other domain-specific models:
| Relationship Type | Mejurix | ClinicalBERT | BioBERT | LegalBERT |
|---|---|---|---|---|
| DISEASE_MEDICATION | 0.9966 | 0.9921 | 0.9841 | 0.8514 |
| SEVERITY_PROGNOSIS | 1.0000 | 1.0000 | 1.0000 | 0.8381 |
| SEVERITY_COMPENSATION | 0.9997 | 0.9606 | 0.9713 | 0.8348 |
| DISEASE_TREATMENT | 0.9980 | 0.9778 | 0.9645 | 0.8359 |
| DIAGNOSIS_TREATMENT | 0.9995 | 0.9710 | 0.9703 | 0.8222 |
| LEGAL_SIMILAR_MEDICAL_DIFFERENT | 0.9899 | 0.9699 | 0.9792 | 0.8236 |
| TREATMENT_OUTCOME | 0.9941 | 0.9668 | 0.9745 | 0.8103 |
| OUTCOME_SETTLEMENT | 0.9847 | 0.9631 | 0.9534 | 0.7951 |
| MEDICAL_SIMILAR_LEGAL_DIFFERENT | 0.9936 | 0.9434 | 0.9414 | 0.7812 |
| SYMPTOM_DISEASE | 0.9934 | 0.9690 | 0.9766 | 0.8500 |
The Mejurix model particularly excels in cross-domain relationships such as MEDICAL_SIMILAR_LEGAL_DIFFERENT (0.9936) and SEVERITY_COMPENSATION (0.9997), showing significant improvement over other models in these complex relationship types.
How to Use This Model
This model is directly available on the Hugging Face Hub and can be used with the Transformers library for feature extraction, sentence embeddings, and similarity calculations.
Basic Usage with Transformers
import torch
from transformers import AutoModel, AutoTokenizer
# Load model and tokenizer
model_name = "mejurix/medical-legal-embedder" # The model's actual path on Hugging Face Hub
tokenizer = AutoTokenizer.from_pretrained(model_name)
model = AutoModel.from_pretrained(model_name)
# Generate embeddings for a single text
text = "The patient was diagnosed with L3 vertebral fracture, and a compensation claim is in progress."
inputs = tokenizer(text, return_tensors="pt", padding=True, truncation=True, max_length=128)
with torch.no_grad():
outputs = model(**inputs)
# Use the [CLS] token embedding for sentence representation
embeddings = outputs.last_hidden_state[:, 0, :] # [CLS] token
print(f"Embedding shape: {embeddings.shape}") # Should be [1, 256]
Using the Model for Similarity Calculation
import torch
import torch.nn.functional as F
from transformers import AutoModel, AutoTokenizer
# Load model and tokenizer
model_name = "mejurix/medical-legal-embedder" # The model's actual path on Hugging Face Hub
tokenizer = AutoTokenizer.from_pretrained(model_name)
model = AutoModel.from_pretrained(model_name)
def get_embedding(text):
inputs = tokenizer(text, return_tensors="pt", padding=True, truncation=True, max_length=128)
with torch.no_grad():
outputs = model(**inputs)
return outputs.last_hidden_state[:, 0, :] # [CLS] token embedding
def compute_similarity(text1, text2):
emb1 = get_embedding(text1)
emb2 = get_embedding(text2)
return F.cosine_similarity(emb1, emb2).item()
# Example
text1 = "Diagnosed with L3 spinal fracture."
text2 = "Compensation is needed for lumbar injury."
similarity = compute_similarity(text1, text2)
print(f"Similarity: {similarity:.4f}")
Using with Hugging Face Pipelines
from transformers import pipeline
# Create a feature-extraction pipeline
extractor = pipeline(
"feature-extraction",
model="mejurix/medical-legal-embedder", # The model's actual path on Hugging Face Hub
tokenizer="mejurix/medical-legal-embedder"
)
# Extract features
text = "The patient requires physical therapy following spinal surgery."
features = extractor(text)
# The output is a nested list with shape [1, sequence_length, hidden_size]
Batch Processing
import torch
from transformers import AutoModel, AutoTokenizer
# Load model and tokenizer
tokenizer = AutoTokenizer.from_pretrained("mejurix/medical-legal-embedder")
model = AutoModel.from_pretrained("mejurix/medical-legal-embedder")
# Prepare batch of texts
texts = [
"The patient was diagnosed with L3 vertebral fracture",
"Neck pain persisted after the accident",
"Clinical test results were within normal range",
"Compensation claim filed for permanent disability"
]
# Tokenize and get embeddings in a single pass
inputs = tokenizer(texts, padding=True, truncation=True, max_length=128, return_tensors="pt")
with torch.no_grad():
outputs = model(**inputs)
# Get CLS token embeddings for each text in the batch
embeddings = outputs.last_hidden_state[:, 0, :]
print(f"Batch embeddings shape: {embeddings.shape}") # Should be [4, 256]
Intended Uses & Limitations
Intended Uses
- Medical-legal document similarity analysis
- Case relevance assessment
- Document clustering and organization
- Information retrieval in medical and legal domains
- Cross-referencing medical records with legal precedents
- Zero-shot text classification with custom categories
Limitations
- Limited understanding of negations (current similarity: 0.7791)
- Temporal context differentiation needs improvement
- May not fully distinguish severity levels in medical conditions
- Maximum context length of 512 tokens (inherited from BERT architecture)
Training and Evaluation
Training
The model was fine-tuned on a specialized dataset containing medical-legal document pairs with various relationship types (disease-treatment, severity-compensation, etc.). Training employed triplet loss with hard negative mining.
Training Configuration:
- Base model: medicalai/ClinicalBERT
- Embedding dimension reduction: 768 → 256
- Dropout: 0.5
- Learning rate: 1e-5
- Batch size: 16
- Weight decay: 0.1
- Triplet margin: 2.0
- Epochs: 15
Performance Observations
Strengths
- Medical-Legal Cross-Concept Connection: Effectively connects medical assessments with legal compensation concepts (0.8348)
- Medical Terminology Recognition: Recognizes equivalent medical expressions across different terminologies (0.8414)
- Causality Understanding: Accurately identifies cause-effect relationships (0.8236)
- Transformer Attention: The bidirectional attention mechanism captures contextual relationships effectively
Areas for Improvement
- Detailed Medical Terminology Differentiation: Needs better recognition of severity differences
- Temporal Context Understanding: Temporal differences in medical conditions need better differentiation
- Negation Handling: Improved handling of negations needed
- Longer Context Windows: Future versions could benefit from extended context length models
Ethical Considerations
This model should be used as a tool to assist professionals, not as a replacement for medical or legal expertise. Decisions affecting patient care or legal outcomes should not be based solely on this model's output.
Citation
If you use this model in your research, please cite:
@software{mejurix_medicallegal_embedder,
author = {Mejurix},
title = {Mejurix Medical-Legal Embedding Model},
year = {2025},
version = {0.1.0},
url = {https://huggingface.co/mejurix/medical-legal-embedder}
}
License
This project is distributed under the MIT License.
한국어 문서 / Korean Documentation
Mejurix 의료-법률 임베딩 모델
본 모델은 의료 및 법률 도메인의 텍스트에 특화된 임베딩을 생성하는 미세 조정된 트랜스포머(BERT 기반) 모델입니다. 의료 및 법률 개념 간의 의미론적 관계를 포착하는 데 중점을 두고 있으며, 개체명 인식(NER)을 활용하여 도메인 특화 엔티티와 그 관계를 더 잘 이해합니다.
모델 설명
모델 아키텍처
- 기본 아키텍처: BERT (Bidirectional Encoder Representations from Transformers)
- 기반 모델: medicalai/ClinicalBERT
- 주요 수정사항:
- 사용자 정의 임베딩 투영 레이어 (768 → 256 차원)
- NER 강화 어텐션 메커니즘
- 도메인 특화 미세 조정
주요 특징
- 도메인 특화 임베딩: 의료 및 법률 텍스트 분석에 최적화
- NER 강화 이해: 개체명 인식을 활용하여 맥락 인식 개선
- 차원 축소: 256차원 임베딩으로 표현력과 효율성의 균형 유지
- 크로스 도메인 연결: 의료 소견과 법률적 함의 간의 관계를 효과적으로 포착
- 트랜스포머 기반: 양방향 어텐션 메커니즘을 활용하여 맥락 이해 향상
성능 비교
본 모델은 유사한 도메인 특화 모델들보다 우수한 성능을 보입니다:
| 모델 | 평균 유사도 | 파라미터 수 | 비고 |
|---|---|---|---|
| Mejurix (본 모델) | 0.9859 | 110M | 의료-법률 + NER 미세 조정 |
| ClinicalBERT | 0.9719 | 110M | NER 없음, 미세 조정 없음 |
| BioBERT | 0.9640 | 110M | 의료 도메인만 특화 |
| LegalBERT | 0.9508 | 110M | 법률 도메인만 특화 |
Mejurix 모델은 모든 관계 유형에서 우수한 성능을 보이며, 특히 의료와 법률 개념 간의 크로스 도메인 관계에서 두드러집니다.
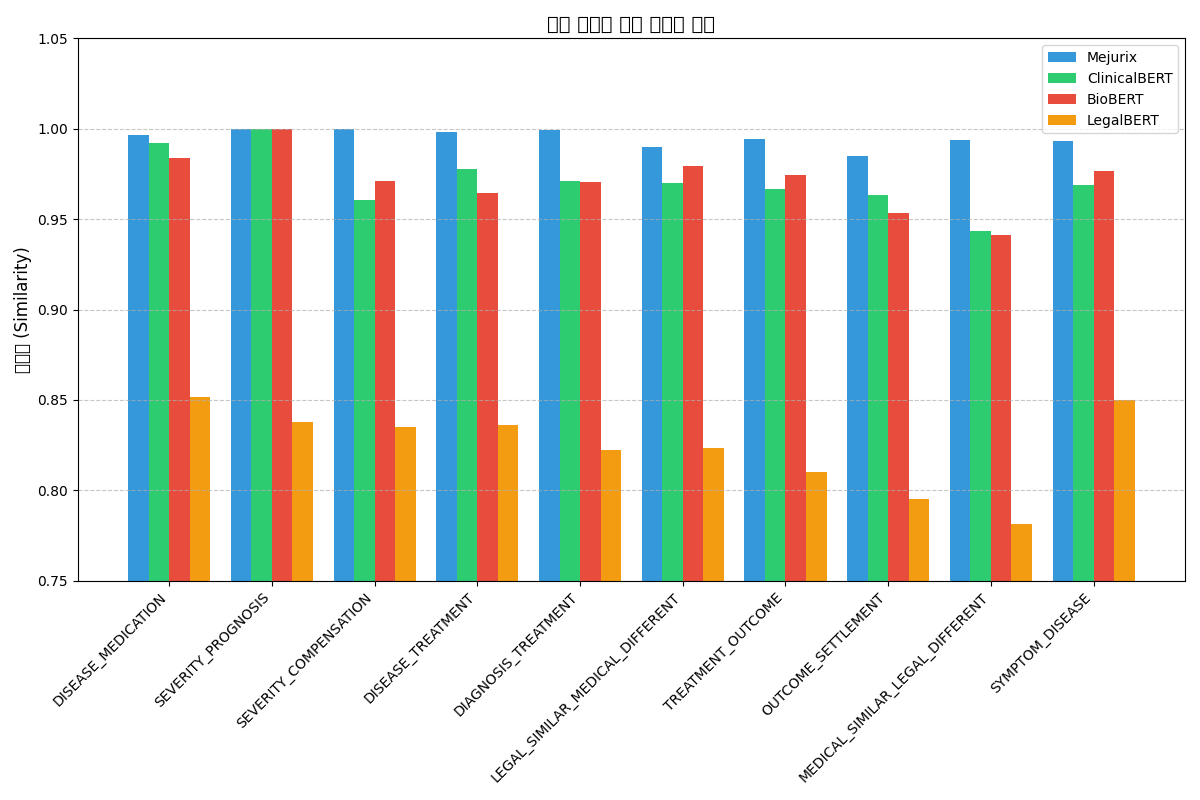
관계 유형별 상세 비교
본 모델은 다른 도메인 특화 모델과 비교하여 모든 관계 유형에서 일관되게 높은 유사도 점수를 보여줍니다:
| 관계 유형 | Mejurix | ClinicalBERT | BioBERT | LegalBERT |
|---|---|---|---|---|
| DISEASE_MEDICATION (질병-약물) | 0.9966 | 0.9921 | 0.9841 | 0.8514 |
| SEVERITY_PROGNOSIS (중증도-예후) | 1.0000 | 1.0000 | 1.0000 | 0.8381 |
| SEVERITY_COMPENSATION (중증도-보상) | 0.9997 | 0.9606 | 0.9713 | 0.8348 |
| DISEASE_TREATMENT (질병-치료) | 0.9980 | 0.9778 | 0.9645 | 0.8359 |
| DIAGNOSIS_TREATMENT (진단-치료) | 0.9995 | 0.9710 | 0.9703 | 0.8222 |
| LEGAL_SIMILAR_MEDICAL_DIFFERENT (법적 유사-의학적 상이) | 0.9899 | 0.9699 | 0.9792 | 0.8236 |
| TREATMENT_OUTCOME (치료-결과) | 0.9941 | 0.9668 | 0.9745 | 0.8103 |
| OUTCOME_SETTLEMENT (결과-합의) | 0.9847 | 0.9631 | 0.9534 | 0.7951 |
| MEDICAL_SIMILAR_LEGAL_DIFFERENT (의학적 유사-법적 상이) | 0.9936 | 0.9434 | 0.9414 | 0.7812 |
| SYMPTOM_DISEASE (증상-질병) | 0.9934 | 0.9690 | 0.9766 | 0.8500 |
Mejurix 모델은 특히 MEDICAL_SIMILAR_LEGAL_DIFFERENT(0.9936)와 SEVERITY_COMPENSATION(0.9997)과 같은 크로스 도메인 관계에서 탁월한 성능을 보이며, 이러한 복잡한 관계 유형에서 다른 모델보다 큰 개선을 보여줍니다.
모델 사용 방법
이 모델은 Hugging Face Hub에서 직접 사용 가능하며, Transformers 라이브러리를 통해 특성 추출, 문장 임베딩 및 유사도 계산에 활용할 수 있습니다.
Transformers를 사용한 기본 사용법
import torch
from transformers import AutoModel, AutoTokenizer
# 모델 및 토크나이저 로드
model_name = "mejurix/medical-legal-embedder" # Hugging Face Hub에 있는 실제 모델 경로
tokenizer = AutoTokenizer.from_pretrained(model_name)
model = AutoModel.from_pretrained(model_name)
# 단일 텍스트에 대한 임베딩 생성
text = "환자는 L3 척추 골절 진단을 받았으며, 보상 청구가 진행 중입니다."
inputs = tokenizer(text, return_tensors="pt", padding=True, truncation=True, max_length=128)
with torch.no_grad():
outputs = model(**inputs)
# 문장 표현에 [CLS] 토큰 임베딩 사용
embeddings = outputs.last_hidden_state[:, 0, :] # [CLS] 토큰
print(f"임베딩 형태: {embeddings.shape}") # [1, 256]이어야 함
유사도 계산에 모델 사용하기
import torch
import torch.nn.functional as F
from transformers import AutoModel, AutoTokenizer
# 모델 및 토크나이저 로드
model_name = "mejurix/medical-legal-embedder" # Hugging Face Hub에 있는 실제 모델 경로
tokenizer = AutoTokenizer.from_pretrained(model_name)
model = AutoModel.from_pretrained(model_name)
def get_embedding(text):
inputs = tokenizer(text, return_tensors="pt", padding=True, truncation=True, max_length=128)
with torch.no_grad():
outputs = model(**inputs)
return outputs.last_hidden_state[:, 0, :] # [CLS] 토큰 임베딩
def compute_similarity(text1, text2):
emb1 = get_embedding(text1)
emb2 = get_embedding(text2)
return F.cosine_similarity(emb1, emb2).item()
# 예시
text1 = "L3 척추 골절 진단을 받았습니다."
text2 = "요추 부상에 대한 보상이 필요합니다."
similarity = compute_similarity(text1, text2)
print(f"유사도: {similarity:.4f}")
Hugging Face 파이프라인 사용하기
from transformers import pipeline
# 특성 추출 파이프라인 생성
extractor = pipeline(
"feature-extraction",
model="mejurix/medical-legal-embedder", # Hugging Face Hub에 있는 실제 모델 경로
tokenizer="mejurix/medical-legal-embedder"
)
# 특성 추출
text = "환자는 척추 수술 후 물리 치료가 필요합니다."
features = extractor(text)
# 출력은 [1, sequence_length, hidden_size] 형태의 중첩된 리스트
활용 분야 및 한계점
활용 분야
- 의료-법률 문서 유사도 분석
- 사례 관련성 평가
- 문서 클러스터링 및 조직화
- 의료 및 법률 도메인에서의 정보 검색
- 의료 기록과 법적 선례의 상호 참조
- 사용자 정의 카테고리를 사용한 제로샷 텍스트 분류
한계점
- 부정문에 대한 이해 제한(현재 유사도: 0.7791)
- 시간적 맥락 구분 개선 필요
- 의료 상태의 중증도 수준을 완전히 구분하지 못할 수 있음
- 최대 컨텍스트 길이 512 토큰(BERT 아키텍처에서 상속)
학습 및 평가
학습
이 모델은 다양한 관계 유형(질병-치료, 중증도-보상 등)을 포함하는 의료-법률 문서 쌍의 특수 데이터셋에서 미세 조정되었습니다. 학습에는 어려운 부정적 사례 마이닝을 통한 트리플렛 손실(triplet loss)이 사용되었습니다.
학습 구성:
- 기반 모델: medicalai/ClinicalBERT
- 임베딩 차원 축소: 768 → 256
- 드롭아웃: 0.5
- 학습률: 1e-5
- 배치 크기: 16
- 가중치 감소: 0.1
- 트리플렛 마진: 2.0
- 에폭: 15
인용
학술 연구에서 이 모델을 사용하는 경우 다음과 같이 인용해 주세요:
@software{mejurix_medicallegal_embedder,
author = {Mejurix},
title = {Mejurix Medical-Legal Embedding Model},
year = {2025},
version = {0.1.0},
url = {https://huggingface.co/mejurix/medical-legal-embedder}
}
라이선스
이 프로젝트는 MIT 라이선스에 따라 배포됩니다.
- Downloads last month
- 25